UTGB Manual
- UTGB Medaka http://medaka.utgenome.org/
Track Information
Welcome to http://utgenome.org/ UTGB (University of Tokyo Genome Browser). With this browser you can browse genomic information of a target species (at present Medaka only) from several aspects. For example, the following tasks can be performed with different combinations.
- You can search gene, by gene name, clone name, cluster name and display its surroundings gene groups.
- You can map a genomic sequence to any genome and observe its surroundings.
- You can save the sequence itself on your system from here.
Questions, suggestions or comments are welcome to annotation at mlab.cb.k.u-tokyo.ac.jp.
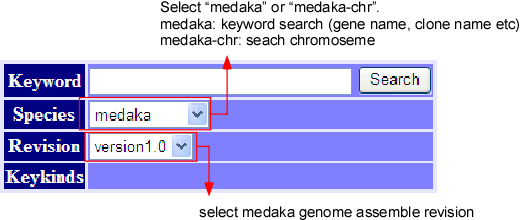
Keyword Search
The top window displays the keyword search engine, as above picture illustrates. For normal keyword search, select "medaka" from species column. To obtain chromosome view (see below), select "medaka-chr" from species column. You can also select the genome assemble revision from revision colomun.
When you select medaka
Inputting any keyword at the query box, we can see the detail information about the query. More about keyword search can be found in the help, written on the same window.
When you select medaka-chr
For Chromosomal view, select species as "medaka-chr" and write chromosome name in keyword then press search button. At present, chromosome name format e.g. "CH01" or "CH16" is only allowed to use.
The details of Chromosomal view is described here: http://medaka3.utgenome.org/~kobayashi/UTGBmedakaHELP/ChromosomeView.html
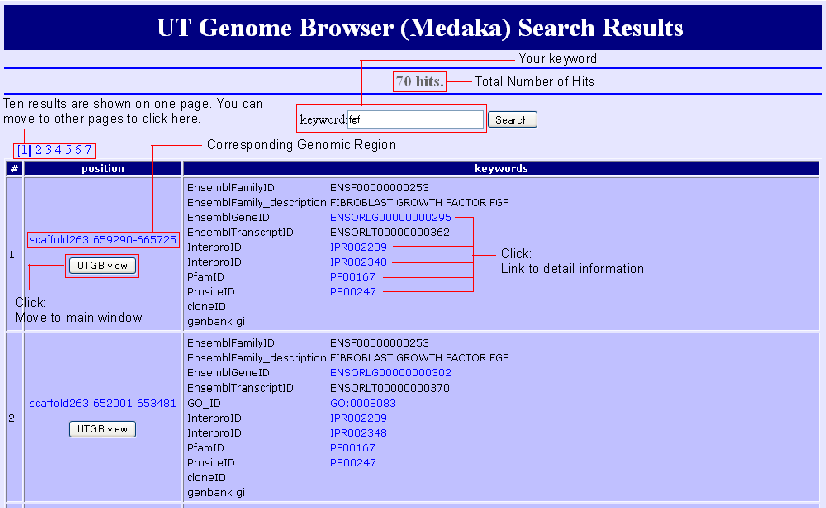
Search Result
Search results are listed by the genomic region because all keywords are connected via regions on the genome. You can move to the mapped region by clicking "UTGBview" button. Hyperlinks are available to access detaled information of some keywords.
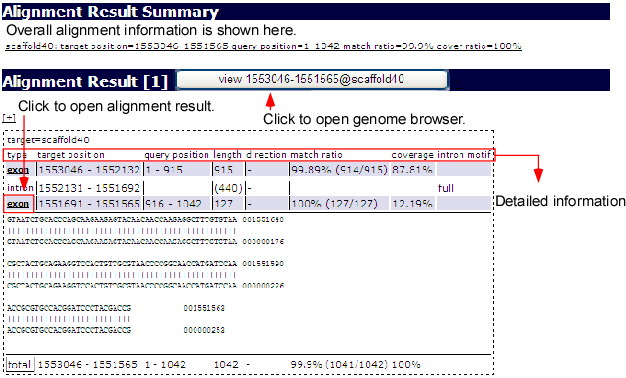
Sequence Alignment
Sequence mapping is possible by inputting a sequence in the online mapping frame (above image) at the bottom of the main window. After selecting the species and its revision, press the search button on the bottom. The request is sent to the alignment server ALPS to align the given sequence to the selected genome. Afterwards, the list of alignment results is returned if the system finds some alignments.
For details in ALPS, see http://ALPS.gi.k.u-tokyo.ac.jp sorry temporary anavailable.
From the list, you can select one alignment by clicking the result button as illustrated above. Then, the aligned result will appear on the browser. The ALPS mapping track in following snapshot displays the added mapping status.
Main Window
The above picture shows the general information about the operation of UT Genome Browser in the main window.
In what follows, we describe the detail about each function.
Scrolling and zooming the main window
On the main window, it is possible to shift and to zoom in/out the present observing sequence. These tasks can be carried out by:
- Inputting value in the input box method.
- Using shifting button method.
- Using overview, ruler and zooming track method.
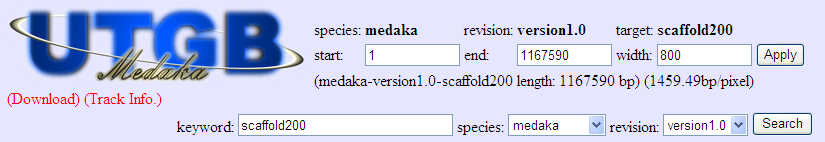
Inputting value in the input box method
You can input values into the input boxes (like above image) inside the main window. The details about this method are as follows,
- Species
- At present, only Medaka is possible to be selected.
- Revision
- Select the revision of genome assembly.
- Start
- Select the start position of the scaffold to display.
- End
- Select the end point of the scsaffold to display. If the atart point is greater than the ending point, the sequence is displayed inversely.
- Width
- Difine the with of the tarcks in pixels here. It is possible to input the width bigger than the screen width. In this case, you can scroll horizontally using scroll bar of the browser.
After manipulating the previous items, press the “Apply” button to apply the condition.
Using shifting button method
Moreover, there is another method by using the button (like above image). Normally, the input box method is useful only when it is important to have the correct position of the sequence. As number must be supplied in this method, sometime this process is quit cumbersome. So we provide more intuitive methods for your convenience. So the buttons related to these methods are as follows,
- Shifting
 ,
,  ,
,  ,
, are buttons to shifting the sequence. The
are buttons to shifting the sequence. The  button scrolls sequence up to one screen size where the
button scrolls sequence up to one screen size where the  button scrolls half screen to the left. The inverse buttons are for shifting to the right.
button scrolls half screen to the left. The inverse buttons are for shifting to the right.
The  button does scaling down and the display will be half of the present size where
button does scaling down and the display will be half of the present size where  button does scaling up and the display will be 2 times bigger than present size. The fractional form button specifies the scale directly. The scaling unit is 1bp/pixel. For example,
button does scaling up and the display will be 2 times bigger than present size. The fractional form button specifies the scale directly. The scaling unit is 1bp/pixel. For example,  means 1 bp is displayed by 1 pixel, whereas
means 1 bp is displayed by 1 pixel, whereas  means 10 bp is displayed by 1 pixel. So
means 10 bp is displayed by 1 pixel. So  means the scale that enlarges the display up to the base level. zoom0001x.png is a scale ratio useful in displaying 1M bps on the screen. At the time of scaling up or down the view points of the present display will be remained unchanged.
means the scale that enlarges the display up to the base level. zoom0001x.png is a scale ratio useful in displaying 1M bps on the screen. At the time of scaling up or down the view points of the present display will be remained unchanged.
Using overview, ruler and zooming track method
In addition, there are some methods to shifting. These are overview, ruler, zooming tracks. You can shift the viewpoint by clicking any position of these tracks. For details, please refer to the respective details about the each track.
Zooming track: http://trac.utgenome.org/project/UTGB/wiki/UTGBMedaka/Track#ZoomingTrack
Overview track: http://trac.utgenome.org/project/UTGB/wiki/UTGBMedaka/Track#OverviewTrack
Ruler track: http://trac.utgenome.org/project/UTGB/wiki/UTGBMedaka/Track#RulerTrack
Functions of buttons
- (rev)
- You can display the sequence in reverse mode by UT Genome Browser. Normally a sequence is displayed from left to right but you can reverse it with the (rev) button. An alternative way to display in reverse way is to input “start” value greater than “end” value in the Input Box. The same task can be done easily by clicking the (rev) button. As the (rev) button reverses start and end points of the present displayed sequence so this can change the display mode from normal to reverse or vice versa. If you press the (fasta) button on reverse mode, then the complementary strand of displayed strand will be retrieved.
- (top)
- By pressing this button, you can return to the top page.
- (track)
- Track prepararion window will appear. Details description will be given below.
- (fasta)
- Present displaying sequence can be retrieved with this button in fasta format. You will be instructed to save as a file name, Medaka-revision-scaffold# (start-end).fasta. e.g. Medaka-version1.0-scaffold12 (345-6789).fasta
- (png)
- Screen shot can be retrieved with this button in PNG format. Click to open the screen shot, and then save the image by right clicking and save. In some browser, the image will be saved as a BMP format.
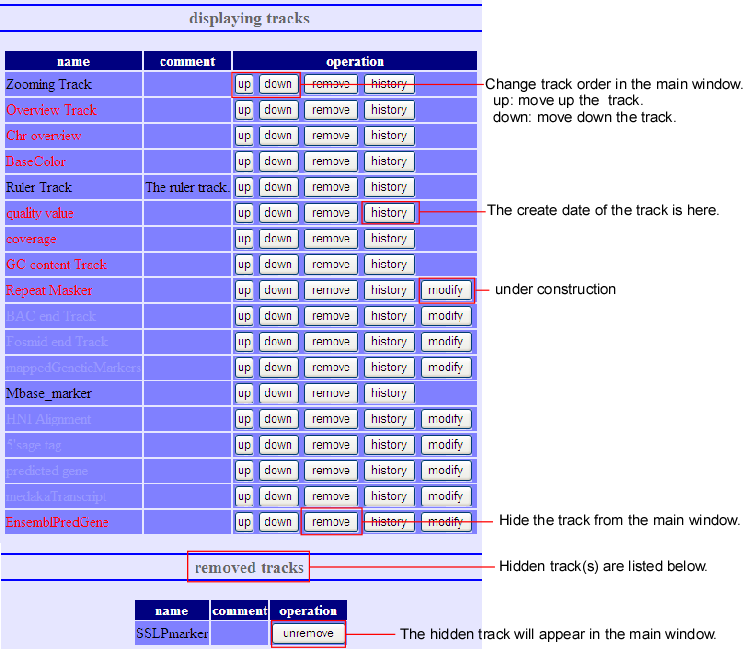
Addition, Deletion, and Rearrangement of Tracks
When the (track) button of the main screen is pressed, the track-editing screen below will pop up. Here you can do addition, deletion and rearrangement of the tracks. This editing screen can be divided into two parts - displaying tracks and removed tracks.
- Displaying track
- Here, tracks’ deletion and rearrangement can be done to the present displayed screen. The deleted tracks will be replaced to next removed track.
- Removed track
- This is the list of all tracks, which are deleted at displaying Tracks. It is something like garbage bin. Undo-processing is possible for the tracks that are displayed here.
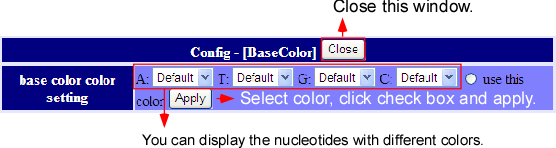
Display configuration
On the main window, when the  button on the left side the track is pressed, the display setting windows will pop up.
There are three parts of setting; style, hide/display and color. The customizing contents of the tracks differ from each other and there are some tracks which contents cannot be customized.
button on the left side the track is pressed, the display setting windows will pop up.
There are three parts of setting; style, hide/display and color. The customizing contents of the tracks differ from each other and there are some tracks which contents cannot be customized.
Default color setting is A:green, G:blue, T:red, C.yellow.
Predicted gene track
5'SAGE track
Medaka Transcripts track
Ensembl predicted gene track
Mutant track
The style select setting is a common function among several tarcks and described below separately.
BAC end tarck and Fosmid end track
Style select setting
There are four kinds of display style. These styles are full, pack, small and dense.
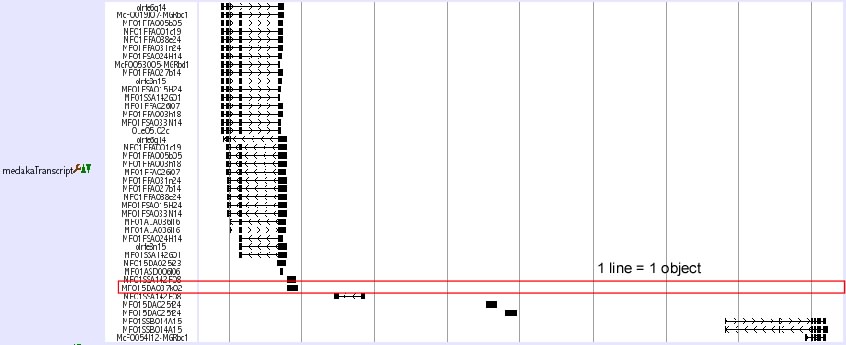
- Full
- In the full style, each object (transcript, BAC clone etc) is displayed in one line. On the left side, Object names sach as clone names are shown.
- Pack
- In the pack style, some objects are shown in the same line. This style can display the tracks more compact than the full style.
- Small
- In this style, object allocation is tha same as the pack style with smaller iages. Object names are not shown in this style.
- Dense
- All objects are stacked in a line.
Ramen Assembler / UT Genome Browser Team Members
Ramen Genome Assembler Development Team
- Development of “Ramen” genome assembler and assembly of medaka genome:
- Masahiro Kasahara and Shin Sasaki
- Development of “Ramen Viewer” for genome assembly:
- Yukinobu Nagayasu
UT Genome Browser Development Team
- Design and development of UT Genome Browser, keyword search function, libraries for describing tracks:
- Yukinobu Nagayasu and Koichiro Doi
- Online mapping function for query sequences:
- Tomoyuki Yamada
- Comparative Genomics Track:
- Yoichiro Nakatani and Wei Qu
- Gene Prediction:
- Ahsan Budrul
- Mapped Gene Track:
- Yasuhiro Kasai
- Database access accelerators:
- Takehiro Furudate and Atsushi Mori
- Overall management:
- Koichiro Doi and Shinichi Morishita
Acknowledgements
This work has been supported by Grant-in-Aid for Scientific Research on Priority Areas (Grant#12209003) to Shinichi Morishita.
Ramen Assembler Development Team members are indebted to Yuji Kohara and Tadasu Shin-i for their technical discussions on the whole genome shotgun assembly.
Members in the UT Genome Browser Development Team are grateful to Kiyoshi Naruse, Daisuke Kobayashi, and Takanori Narita for their valuable input to improve the functions of the browser in a variety of ways.
Attachments (63)
- browser_main.png (34.5 KB) - added by 18 years ago.
- browser_moves.png (3.4 KB) - added by 18 years ago.
- browser_params.png (5.8 KB) - added by 18 years ago.
- browser_track.png (23.0 KB) - added by 18 years ago.
- cfg.png (715 bytes) - added by 18 years ago.
- f.png (723 bytes) - added by 18 years ago.
- ff.png (763 bytes) - added by 18 years ago.
- image001.jpg (13.5 KB) - added by 18 years ago.
- keyword_genes.png (15.0 KB) - added by 18 years ago.
- keyword_locs.png (17.9 KB) - added by 18 years ago.
- keyword_top.png (13.5 KB) - added by 18 years ago.
- online01.PNG (9.3 KB) - added by 18 years ago.
- online02.PNG (7.0 KB) - added by 18 years ago.
- online02.2.PNG (7.0 KB) - added by 18 years ago.
- online03.PNG (28.4 KB) - added by 18 years ago.
- r.png (730 bytes) - added by 18 years ago.
- rr.png (758 bytes) - added by 18 years ago.
- track_overview.png (3.3 KB) - added by 18 years ago.
- zoom0001x.png (802 bytes) - added by 18 years ago.
- zoom001x.png (801 bytes) - added by 18 years ago.
- zoom01x.png (787 bytes) - added by 18 years ago.
- zoom1x.png (779 bytes) - added by 18 years ago.
- zoom10x.png (786 bytes) - added by 18 years ago.
- zoomin.png (760 bytes) - added by 18 years ago.
- zoomout.png (759 bytes) - added by 18 years ago.
- rr.2.png (758 bytes) - added by 18 years ago.
- genescan.jpg (4.8 KB) - added by 18 years ago.
- comp.jpg (20.1 KB) - added by 18 years ago.
- image029.jpg (2.2 KB) - added by 18 years ago.
- image029.2.jpg (2.2 KB) - added by 18 years ago.
- image030.jpg (2.1 KB) - added by 18 years ago.
- image030.2.jpg (2.1 KB) - added by 18 years ago.
- image032.jpg (3.0 KB) - added by 18 years ago.
- image033.jpg (3.0 KB) - added by 18 years ago.
- image033.2.jpg (3.0 KB) - added by 18 years ago.
- image034.jpg (976 bytes) - added by 18 years ago.
- image035.jpg (3.5 KB) - added by 18 years ago.
- image036.jpg (5.9 KB) - added by 18 years ago.
- image037.jpg (29.4 KB) - added by 18 years ago.
- image038.jpg (7.0 KB) - added by 18 years ago.
- image039.jpg (4.8 KB) - added by 18 years ago.
- image040.jpg (1.8 KB) - added by 18 years ago.
- image041.jpg (1.5 KB) - added by 18 years ago.
- image041.2.jpg (1.5 KB) - added by 18 years ago.
- Main_Page.png (93.8 KB) - added by 18 years ago.
- Operation.png (54.1 KB) - added by 18 years ago.
- function_button.png (1.6 KB) - added by 18 years ago.
- align_result.png (21.1 KB) - added by 18 years ago.
- Online_mapping.png (13.8 KB) - added by 18 years ago.
- Online_mapping2.png (13.8 KB) - added by 18 years ago.
- ALPS.png (6.4 KB) - added by 18 years ago.
- keyword_top2.png (11.0 KB) - added by 18 years ago.
- browser_track2.png (62.5 KB) - added by 18 years ago.
- browser_track3.png (62.5 KB) - added by 18 years ago.
- BAC_Fosmid_config.png (33.4 KB) - added by 18 years ago.
- Basecolor_config.png (10.7 KB) - added by 18 years ago.
- full.jpg (48.3 KB) - added by 18 years ago.
- small.jpg (16.3 KB) - added by 18 years ago.
- pack.jpg (41.3 KB) - added by 18 years ago.
- dense.jpg (8.6 KB) - added by 18 years ago.
- Style_setting.jpg (11.9 KB) - added by 18 years ago.
- Search_Result.png (17.6 KB) - added by 18 years ago.
- Search_Result2.png (17.6 KB) - added by 18 years ago.
Download all attachments as: .zip